The parameter map of a partial differen
The intriguing thing here is that this is a single chemical system! It doesn't fit into the usual characterisation of activator and inhibitor, like Gray-Scott or Turing's original model.
The formula is:
du/dt = mu*u - bilaplacian(u) - 2*laplacian(u) - u + beta*u*u - u*u*u
In this parameter map, mu ranges from -1 on the left to 5 on the right, and beta ranges from -3 at the bottom to 3 at the top.
The bilaplacian is the laplacian operator applied twice. In a one-dimensional system if your laplacian is computed by a finite differencing kernel of 1,-2,1 then the bilaplacian kernel is 1,-4,6,-4,1.
The formula comes from the work of +Matt Pennybacker:
http://pennybacker.net
(Matt suggested trying it without the gradient term in the paper.)
There are some Ready files here to explore:
http://code.google.com/p/reaction-diffusion/source/browse/trunk/Ready#Ready%2FPatterns%2FPennybacker2013
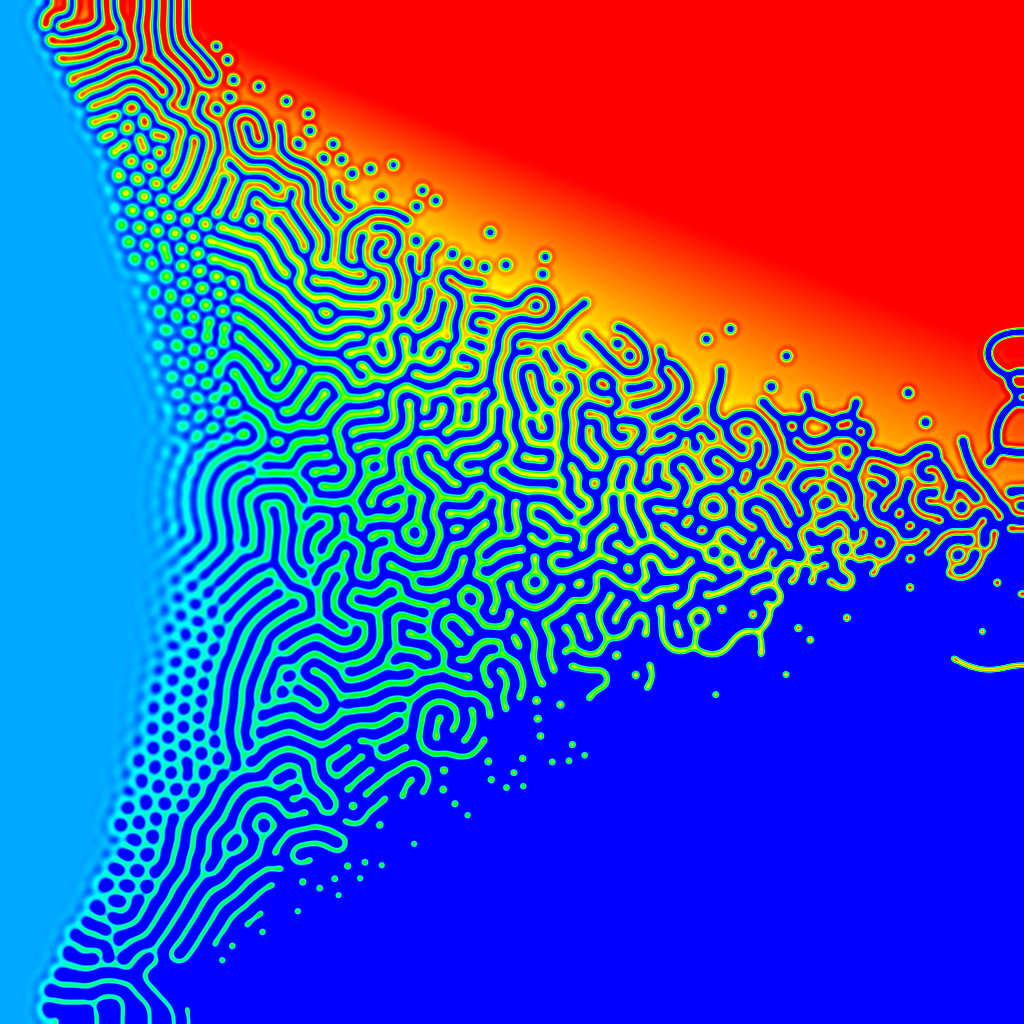
The parameter map of a partial differential equation, showing a reasonably standard pattern of Turing instabilities for different combinations of parameters - spots and labyrinthine stripes. The intriguing thing here is that this is a single chemical system! It doesn't fit into the usual characterisation of activator and inhibitor, like Gray-Scott or Turing's original model. The formula is: du/dt = mu*u - bilaplacian(u) - 2*laplacian(u) - u + beta*u*u - u*u*u In this parameter map, mu ranges from -1 on the left to 1 on the right, and beta ranges from -3 at the bottom to 3 at the top. The bilaplacian is the laplacian operator applied twice. In a one-dimensional system if your laplacian is computed by a finite differencing kernel of 1,-2,1 then the bilaplacian kernel is 1,-4,6,-4,1. The formula comes from the work of +Matt Pennybacker: http://pennybacker.net (Matt suggested trying it without the gradient term in the paper.) There are some Ready files here to explore: http://code.google.com/p/reaction-diffusion/source/browse/trunk/Ready#Ready%2FPatterns%2FPennybacker2013
This post was originally on Google+

(See page 10). I wonder if it is at all related.